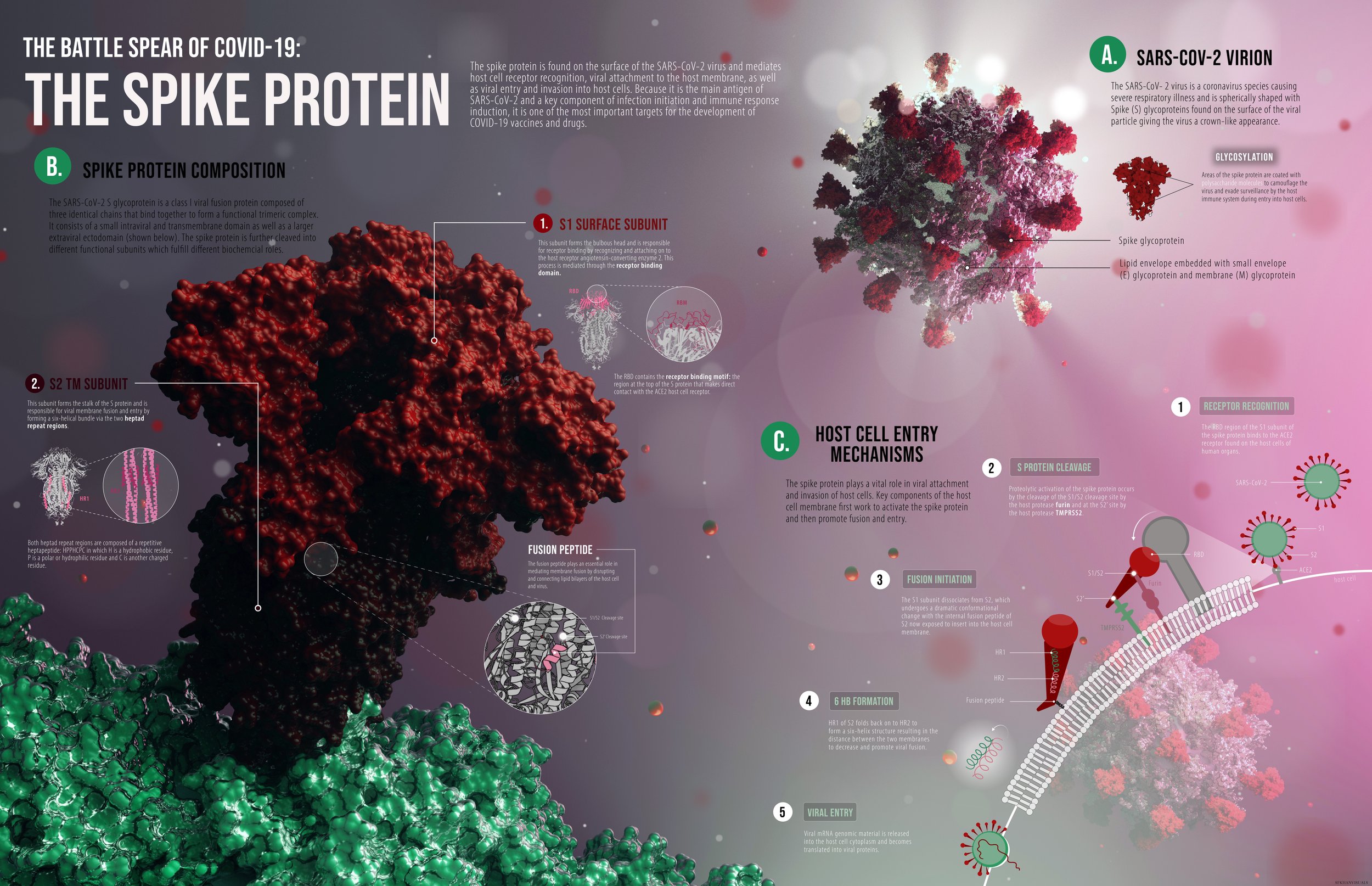
The Spike Protein.
Date: July 15, 2021
Medium: Autodesk Maya, Adobe Illustrator
Audience: Educated Lay audience
Purpose: To visualize the molecular structure and function of a biologically relevant macromolecule. I chose to illustrate the components and the mechanism of action of the SARS- COV-2 virus spike protein.
DEVELOPMENT 1: SKETCHING
A very rough sketch of the layout; on the left is a closeup for the spike protein sitting on the virus, and on the right is this protein’s MOA in allowing the virus to enter a host cell (left). The components of this draft were processed in 3D protein imager and brought into Maya to plan arrangement of 3d models. One of the issues I faced was highlighting the S protein cleavage sites which are embedded deep in the protein and hidden from view (right).
DEVELOPMENT 2: COLOUR THUMBNAILS
Quick and rough colour thumbnails were made to explore colour palettes on top of a basic layout sketch.
DEVELOPMENT 3: DATA EXTRACTION
Structural data of the spike protein was extracted from the Protein data bank. PDB data was brought into 3D protein imager where glycosylation sites on the spike protein were made more distinct.
DEVELOPMENT 4: FINAL RENDERING
3D RENDER: The spike protein was further processed in VMD and chimera to separate out the S1 and S2 subunit. It was brought into Maya and I created a scene of a molecular space which I then rendered to create the background for my visualization. I decided to add a third section visualizing the host cell entry mechanisms of the spike protein.









